
Biosynthesis Captured in Motion
By:
- Susan Brown
Published Date
By:
- Susan Brown
Share This:
Article Content
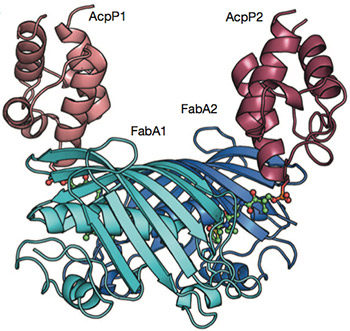
Linking carrier proteins (red) to enzymes (blue) that synthesize fatty acids, reveals this snapshot of biosynthesis in action.
Chemists have caught molecules in the act of biosynthesis revealing an animated view of how a fundamental piece of cellular machinery operates. The system they observed, a critical metabolic pathway, generates fatty acids, essential components of fats and structures such as cell membranes. Nature published their findings in the early online edition December 22.
Scientists would like to regulate this pathway for diverse purposes: to curb the chaotic growth of cancer cells or harmful bacteria, for example, or to boost the production of oils by algae to be harvested for fuel. But these efforts often fail, for lack of complete information about how molecules involved in the synthesis interact.
“We need to decipher the communication code, to understand how proteins work with each other on a molecular level,” said Michael Burkart, a biological chemist at the University of California, San Diego, who led the project in partnership with Shiou-Chuan (Sheryl) Tsai, a biochemist at UC Irvine.
The challenge is that biological molecules at work are usually in motion. “It’s analogous to a huge machine,” Tsai said. “We need to know how different parts of the engine fit and work together.”
Capturing images of the varying configurations required a team of chemists with complementary areas of expertise. Their joint work has revealed the molecular dance of a carrier protein that protects the growing chain of fatty acids and ferries it between active sites of the enzymes that assemble it.
Tsai is an expert in x-ray crystallography, a method chemists use to lock proteins in place and generate still images of their structures. The protein this team studied, acyl carrier protein or ACP, proved particularly challenging because it moves so much.
To pin down ACP and other proteins, Burkart’s research group has developed a molecular toolkit, a set of small molecules that can lock proteins together in their working configurations.
“Mike pins them, and I take the pictures,” said Tsai, who holds appointments in the departments of Molecular Biology and Biochemistry, Chemistry, and Pharmaceutical Sciences at UC Irvine. The strategy resulted in two snapshots, showing two different states of interaction.
Then, working with Stanley Opella, professor of chemistry and biochemistry at UC San Diego, they extended their observation using a different method, nuclear magnetic resonance, which reveals the motions of the carrier protein and enzyme in solution. These observations were validated by dynamic modeling of the molecule by a research group led by J. Andrew McCammon, also a professor of chemistry and biochemistry at UC San Diego.
They saw salt bridges form and the enzyme grab helices and pull them apart. The growing chain of fatty acids also loops back into a pocket in the carrier molecule. “We call it the marsupial protein,” Burkart says. “It protects its young.” That is, until it’s time for the product to go.
“We thought this tiny protein was just a transporter,” Tsai said. “But when it ejects its cargo, it works like a piston. We see a helical collapse forcing the fatty acid out.”
The approach will allow this team to continue to unravel interactions between proteins, a critical step to the successful manipulation of biosynthetic pathways.
Additional authors include Chi Nguyen, Joel Bruegger, Grace Caldara-Festin and David Jackson of UC Irvine; and Robert Haushalter, John Lee, Phineas Markwick, Kara Finzel, Fumihiro Ishikawa and Bing O’Dowd of UC San Diego. NIGMS supported this work with grants to Burkart and Tsai (GM 100305 and GM095970). Part of the work was carried out at the Stanford Synchotron Radiation Lightsource, which is supported by the U.S. Department of Energy.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.


