
Exploring Evolutionary Relationships through CIPRES
SDSC Gateway Provides Advanced Software and HPC Tools Needed by Today’s Biologists
Published Date
By:
- Warren Froelich
Share This:
Article Content
For researchers and others interested in the genetic relationships of our planet’s living creatures, the research was groundbreaking, described as a “momentous discovery” that advanced the most important organizing principle of biology: the “tree of life.”
For Laura Hug, then a postdoctoral fellow at UC Berkeley and a co-author of the paper published in the April 11, 2016 edition of Nature Microbiology, the research also was a crowning achievement, a career-changer.
“This paper has had the largest impact of any in my career to date,” said Hug, now an assistant professor and Canada Research Chair in Environmental Microbiology at the University of Waterloo. “And it would not have been possible without CIPRES.”
CIPRES, for CyberInfrastructure for Phylogenetic RESearch, is a web-based portal or “gateway” launched in 2009 at the San Diego Supercomputer Center (SDSC) at UC San Diego that allows researchers to explore evolutionary connections among species using supercomputers provided by the National Science Foundation’s (NSF’s) XSEDE (eXtreme Science and Engineering Discovery Environment) project.
The gateway provides access to a sophisticated set of software tools and high-performance computers that would be both costly and difficult for individual researchers to create. It allows phylogenetic researchers to compare DNA sequences and predict common ancestors among plants and animals quickly and efficiently, providing them with a workspace where they can organize and repeat their analyses, and store their results indefinitely.
“Phylogenetic analyses are critical since the evolutionary origin of the DNA, protein or life-form under study is a fundamental aspect of nearly every problem in modern biology, whether it be the spread of a virus, or understanding the origin of particular protein,” said Mark Miller, the gateway’s principal investigator based at SDSC. “Rapid advances in DNA sequencing has produced a deluge of useful DNA sequence data, so it is possible to conduct ever-more sensitive analyses, and ask bigger questions.”
However, the increase in DNA data means the computational power required for routine phylogenetic analyses is increasing exponentially. Moreover, advances in sequence alignment and tree interference algorithms continue. CIPRES provides access to the compute resources needed and access to up-to-date versions of complex software packages. Researchers without access to the computational tools provided by CIPRES are at a significant disadvantage.
Origins and Evolution of CIPRES
“CIPRES grew organically out of an award from the NSF, from a program in 2003 called Information Technology Research,” Miller recalled. “The intent of the grant was to create radical new tools to solve problems that will appear in the future, but researchers in the field made it clear they wanted help sooner rather than later. So the project leadership decided to create a web resource to meet that need.”
By the close of the initial award, CIPRES had evolved into a gateway linked to supercomputing clusters available through the NSF-funded TeraGrid (now XSEDE), which provides academic researchers with the most advanced collection of integrated digital resources and services in the world.
With the pent-up demand for such a service, researchers – almost akin to a “Black Friday” moment – lined up to gain access to the new gateway. With each passing year, the demand has continued to grow.
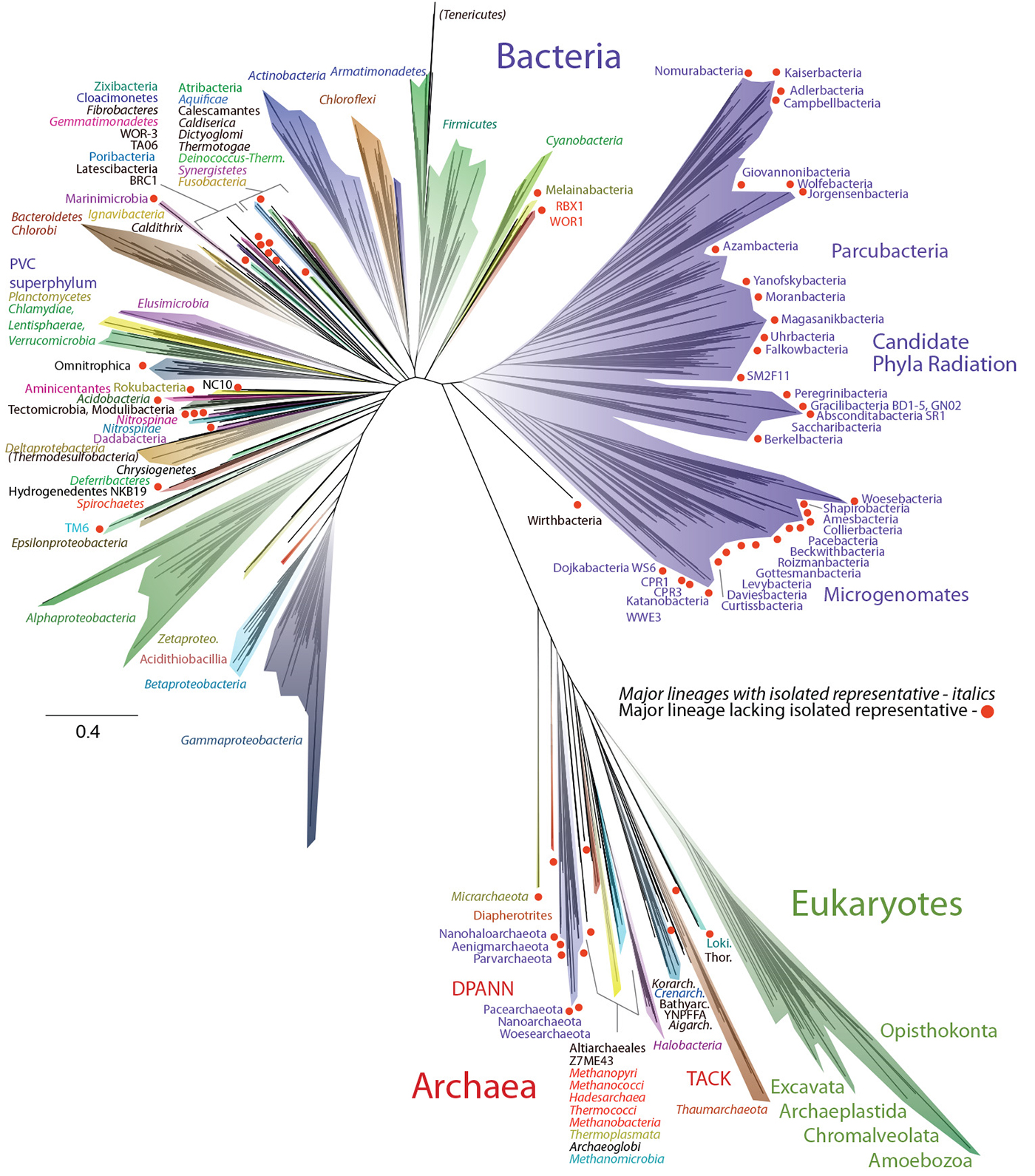
An image of the new ‘Tree of Life’. Courtesy of Laura Hug, Jill Banfield, and Nature Microbiology.
“Looking back, I wasn’t surprised at all by the demand,” said Miller. “People were working mostly on their laptops, so imagine making a run where your laptop becomes unresponsive for a month, or imagine if it gets closed or unplugged mid-run, and all your investment is lost. We focused strictly on providing access to the things users couldn’t do for themselves, which is access to phylogenetic codes on high-performance computing (HPC) resources.”
Added Nancy Wilkins-Diehr, SDSC associate director, principal investigator for the NSF-funded Science Gateway Community Institute, and co-PI of XSEDE: “I believe the success of CIPRES is due to the extremely high level of customer service provided by the CIPRES team. They’ve deployed top quality tools of interest to the community, optimized and installed on supercomputers. And they’ve put in the extra effort to really understand their community.”
Lasting Impact, Global Appeal
The results, in terms of scientific impact, have been dramatic.
- Since its launch about eight years ago, more than 20,000 authenticated users have run one or more jobs using CIPRES, from 86 countries around the world. In the past year, more than 4900 new users accessed HPC resources for the first time through CIPRES.
- In March 2016, some 92 percent of users responding to an annual survey said access to HPC provided by CIPRES benefited their research program in a tangible way, while 83 percent said CIPRES allowed them to accomplish something that would have been difficult or impossible without this resource.
- At least 3,500 peer-reviewed publications in journals have relied on CIPRES resources for their research, in a wide range of biological fields.
Brent Mishler, a professor of integrative biology at UC Berkeley and one of the co-PIs on the original CIPRES NSF grant, has used the resource to develop an entirely new field of research which he calls “spatial phylogenetics”, the goal of which is to develop quantitative measures of biodiversity and endemism -- the ecological state of a species being unique to a defined geographic location, such as an island, nation, country or other defined zone, or habitat type.
“As a community-built resource, CIPRES addresses what the scientists really want and need to do in the real world of research,” said Mishler.
Aside from increasing our understanding of the evolutionary relationships of this planet’s diverse range of species, the research also has yielded results of critical importance to the health and welfare of humans.
- One study published in Science described how African sleeping sickness, which continues to plague the African continent with human and economic casualties, deceives the body’s immune system, opening a key step toward eradicating the disease.
- Other research, published in Nature Communications, identified mutations in highly lethal strains of bird flu, showing how swine flu may also carry similar mutations.
- A research group at SUNY Buffalo found that genetic elements of filoviruses, including often-fatal hemorrhagic diseases such as Ebola and Marburg, are integrated into the genomes of bats, rodents, shrews, and other small mammals, providing evidence that these animals serve as reservoirs for human infection.
- And the list goes on, including new information about the transmission and virulence of human hanta, influenza and HIV viruses.
Indeed, many researchers have offered tributes about how CIPRES has launched their careers or otherwise allowed them to finish research that otherwise seemed intractable with their resources.
“We’ve never marketed this resource; it just took off by word of mouth,” said Miller. “Every now and then someone will write me an unsolicited note to say how much they appreciate the service we provide. That always feels good.”
Some testimonials come from off-the-beaten path.
“I’m in the jungles of Panama for fieldwork,” said Jesse Delia, a researcher from UC Berkeley. “I can’t tell you how helpful it is to have this online resource.”
Another from Rick Miller, a biologist from Southeastern Louisiana University: “I am a faculty member at a small regional university; we have limited computing facility and nothing like the computing power available at R1 (research) universities. Without the amazing resources available through CIPRES, my research would have been crippled.”
Others come from young researchers seeking a legacy for their first big discovery.
“For young researchers at the transition of postdoc to PI, CIPRES is an important resource because we usually lack (our) own funding/infrastructure for these kind of analyses,” said Alexander Suh, an assistant professor in the Department of Evolutionary Biology, Uppsala University, Sweden.
Suh was part of an international team that found DNA “fossils” of parasitic nematodes left several millions of years ago in seven groups of birds, a study that has implications for how genes jump from species to create some human diseases.
“CIPRES has helped me a lot toward achieving scientific independence,” Suh said.
Laura Hug, now with her own lab at Waterloo University, says she is a big CIPRES fan. She recalls how concerned that she would not be able to finish her work advancing the “tree of life”, due to a lack of computational power needed to conduct the “phylogenetic inference” at the heart of her research.
“I ran into insufficient memory, hard time-caps on jobs, and other reasons for jobs failing,” she recalled. “After several months, I had not completed a single iteration of the tree! I was extremely frustrated.”
Then, through a Google search, she found CIPRES.
“Within two to three working days, I was able infer phylogeny, allowing me to iterate through several additions and adjustments to the dataset in question, leading to the final publication,” she said. “I was extremely relieved to have found a solution to my problem.”
The new tree placed a spotlight on bacteria and Archaea lurking in the Earth’s nooks and crannies, providing proof that the life we see around us – plants, animals, humans and other so-called eukaryotes – represent but a tiny fraction of the world’s biodiversity. And it grabbed international headlines in some of the world’s most prestigious science publications and newspapers.
Quoted in the New York Times, Brian P. Hedlung, a microbiologist from the University of Las Vegas who was not involved in the research, offered his analysis of the study: “Most of life is hiding under our noses.”
Today, Hug says she routinely uses CIPRES for phylogenetic inference in her new lab, and she anticipates using the gateway to maximize her computational capacity in the future.
“I’ve been advertising CIPRES in nearly every talk I’ve given in the past two years,” she added.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.


