
RCSB Protein Data Bank Launches Mobile Application
SDSC Helps Develop Free ‘App’ to Access 100,000+ Molecule Structures
By:
- Jan Zverina
Media Contact:
- Jan Zverina - jzverina@sdsc.edu
- Warren R. Froelich - froelich@sdsc.edu
Published Date
By:
- Jan Zverina
Share This:
Article Content
The RCSB Protein Data Bank (PDB), which recently archived its 100,000th molecule structure, has introduced a free mobile application device that enables users from the general public and expert researchers to quickly search and visualize the 3D shapes of proteins, nucleic acids, and molecular machines.
RCSB PDB Mobile is freely available from the Apple App Store and Google Play. More information is at www.rcsb.org.
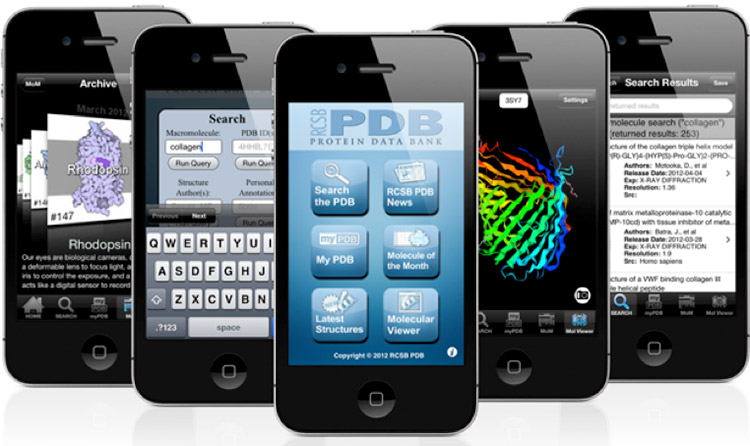
RCSB Protein Data Bank
“As the mobile web is starting to surpass desktop and laptop usage, scientists and educators are beginning to integrate mobile devices into their research and teaching,” said Peter Rose, a researcher with the San Diego Supercomputer Center (SDSC) at the University of California, San Diego, and Scientific Lead with the RCSB PDB. “In response, we have developed this application for iOS and Android mobile platforms to enable fast and convenient access to RCSB PDB data and services.”
RCSB PDB is a vital resource for biological research and education worldwide that provides enhanced access to information about the 3D structures of nucleic acids, proteins, and large molecular machines contained in the Protein Data Bank (PDB) archive. Located at Rutgers, The State University of New Jersey and the UC San Diego’s SDSC/Skaggs School of Pharmacy and Pharmaceutical Sciences at UC San Diego, RCSB PDB develops query, visualization, and analysis tools that help researchers understand many facets of biology and medicine.
According to a paper published online earlier this month on the Oxford Journal’s Bioinformatics website, the goal was to produce an intuitive app with the following core capabilities: a simple search interface, quick browsing of search results, a view of basic data about a structure entry and its Pub-Med abstract, and high-performance molecular visualization. In addition, the app provides access to the RCSB PDB Molecule of the Month educational series, and can be used to store personal notes and annotations.
“Analysis of RCSB PDB website statistics when we initiated development efforts showed that iOS access far outstripped Android site visits,” according to the paper. “Moreover, our experience with development tools and API suggested that iOS would to be a significantly more robust system, with fewer device form factors to support and less OS fragmentation. Therefore, iOS was chosen as the first development project, followed by development of the native Android app.”
In addition to SDSC’s Rose, researchers involved in developing the RCSB PDB Mobile application include Gregory Quinn, Chunxiao Bi, Cole Christie, Kyle Pang, and Andreas Prlić from SDSC and the RCSB PDB; Takanori Nakane from the Graduate School of Medicine, Kyoto University; Christine Zardecki, Maria Voigt, and Helen Berman from the RCSB PDB and Rutgers University in New Jersey; and Philip Bourne, formerly with UC San Diego’s Skaggs School of Pharmacy and Pharmaceutical Sciences and now with The National Institutes of Health in Bethesda, Maryland.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.


