
SDSC’s Gordon Supercomputer: Parsing Genes, Proteins, and Big Bio Data
By:
- Cassie Ferguson
Published Date
By:
- Cassie Ferguson
Share This:
Article Content
Gordon, the newest high-performance supercomputer at the San Diego Supercomputer Center (SDSC) at the University of San Diego, California, has proven to be a boon to biologists interested in rapidly sifting through an ever-expanding amount of data.
“Next-generation sequencing has profoundly transformed biology and medicine, providing insight into our origins and diseases,” according to Wayne Pfeiffer, a Distinguished Scientist at SDSC. “However, obtaining that insight from the data deluge requires complex software and increasingly powerful computers.”
Related Story
This story is the second in a series highlighting projects using the San Diego Supercomputer Center’s newest HPC resource. Read part one here.
Available for use by industry and government agencies, Gordon also is part of the NSF’s XSEDE (eXtreme Science and Engineering Discovery Environment) program, a nationwide partnership comprising 16 supercomputers as well as high-end visualization and data analysis resources. Full details on Gordon can be found at http://www.sdsc.edu/supercomputing/gordon/
Following are three examples of how Gordon’s unique features – such as its large-scale deployment of flash storage – have improved the scope and accessibility of essential biological research databases, and have optimized novel work connecting social networks and genetics.
One Search to Bind Them: IntegromeDB
The diversity of biological fields has spawned thousands databases and millions of public biomedical, biochemical, and drug and disease-related resources. For researchers interested in collecting information from those resources, search engines such as Google are of limited use since they are unable to comprehend the language of biology. They return results on the basis of keywords rather than in terms of scientific importance.
Michael Baitaluk and Julia Ponomarenko, Principal Investigators at SDSC, created a “smart” search for biologists, one able to return gene- and protein-centered knowledge in a biologically meaningful way – for example, by pathways, binding partners, structures, mutations, associated diseases, splice variants, or experiments. Called IntegromeDB for its ability to integrate biomedical data, the resource includes more than 16 million experimental findings receptor binding data for drugs and bioactive compounds, kinetic information for drug-metabolizing enzymes, and relevant signaling proteins are semantically linked to nearly 120 ontologies with a controlled vocabulary of approximately 70 million synonyms. Since its launch in January 2012, more than 4,000 users have visited the resource and it has become an official Science Gateway for the National Science Foundation.
Stored in a PostgreSQL database, IntegromeDB contains over 5,000 tables, 500 billion rows, and 50 terabytes of data. Baitaluk predicts IntegromeDB will eventually require more than a single petabyte of storage. The resource utilizes sixteen compute and four I/O nodes of Gordon and 150 terabytes on SDSC’s Data Oasis.
Unclogging a Bottleneck for the Protein Data Bank
Nearly 250,000 scientists take advantage of the RCSB Protein Data Bank each month, every one of them depending on the resource to quickly provide details on over 90,000 proteins, nucleic acids, and complex assemblies. While the PDB’s current resources can easily handle research requests such as pairwise protein comparisons, the calculation of large numbers of protein structure alignments is too computationally intensive to be done in real time. So the PDB pre-calculates a large number of pairwise three-dimensional protein structure alignments and makes them available via its website.
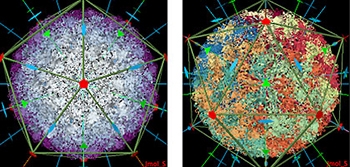
Scientists using the PDB can examine the symmetry and stoichiometry of structures such as these. In these images the virus causing Foot-and Mouth Disease has icosahedral symmetry. Image by Andreas Prlic, UC San Diego
Periodically, those alignments are recalculated as new protein structures are deposited into the database. However, the process of updating slows at the server that stands between the PDB and the nodes used to perform the alignment calculations. Much like an overwhelmed traffic interchange, the system cannot keep up with the data going and coming from the database, creating a data traffic jam. Phil Bourne, professor of pharmacology at UC San Diego, and Andreas Prlic, a senior scientist with the university, investigated whether this process could be improved using Gordon.
They found that the calculations were sped up 3.8 times, the previous calculations that required 24 hours now taking 6.3 hours. To gauge whether I/O performance might improve even further, Bourne and his colleagues tried the same calculations using Intel’s new Taylorsville flash drives. The drives deliver twice as much bandwidth and read IOPS, and 13 times more write IOPS than Intel’s Lyndonville flash drives. This drove the time down to 4.1 hours.
“With its excellent communications capabilities, Gordon can be used to greatly reduce the time to solution over the systems we currently use,” said Bourne.
Making Genome-wide Connections
James Fowler, professor of medical genetics and political science at UC San Diego, and Nicholas Christakis, professor of sociology, medicine, and health care policy at Harvard University, have made news with their ability to connect social networks to the spread of disease such as obesity. Their headline-grabbing research is backed up with a series of computational steps of data preparation, statistical inferencing, and quality checks.
Each step is often performed by specialized software, some of it requiring intense computation to process the DNA of hundreds of thousands of people and millions of pieces of gene sequence. According to Fowler, his analyses can be limited by hardware, making it necessary to use a resource such as Gordon.
“Gordon with the Lustre file system provided an efficient environment where these jobs could be set up, data could be split up or combined, and variations on sampling and variance estimated procedures could be tested,” said Fowler.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.


