
Viral Switches Share a Shape
Published Date
By:
- Susan Brown
Share This:
Article Content
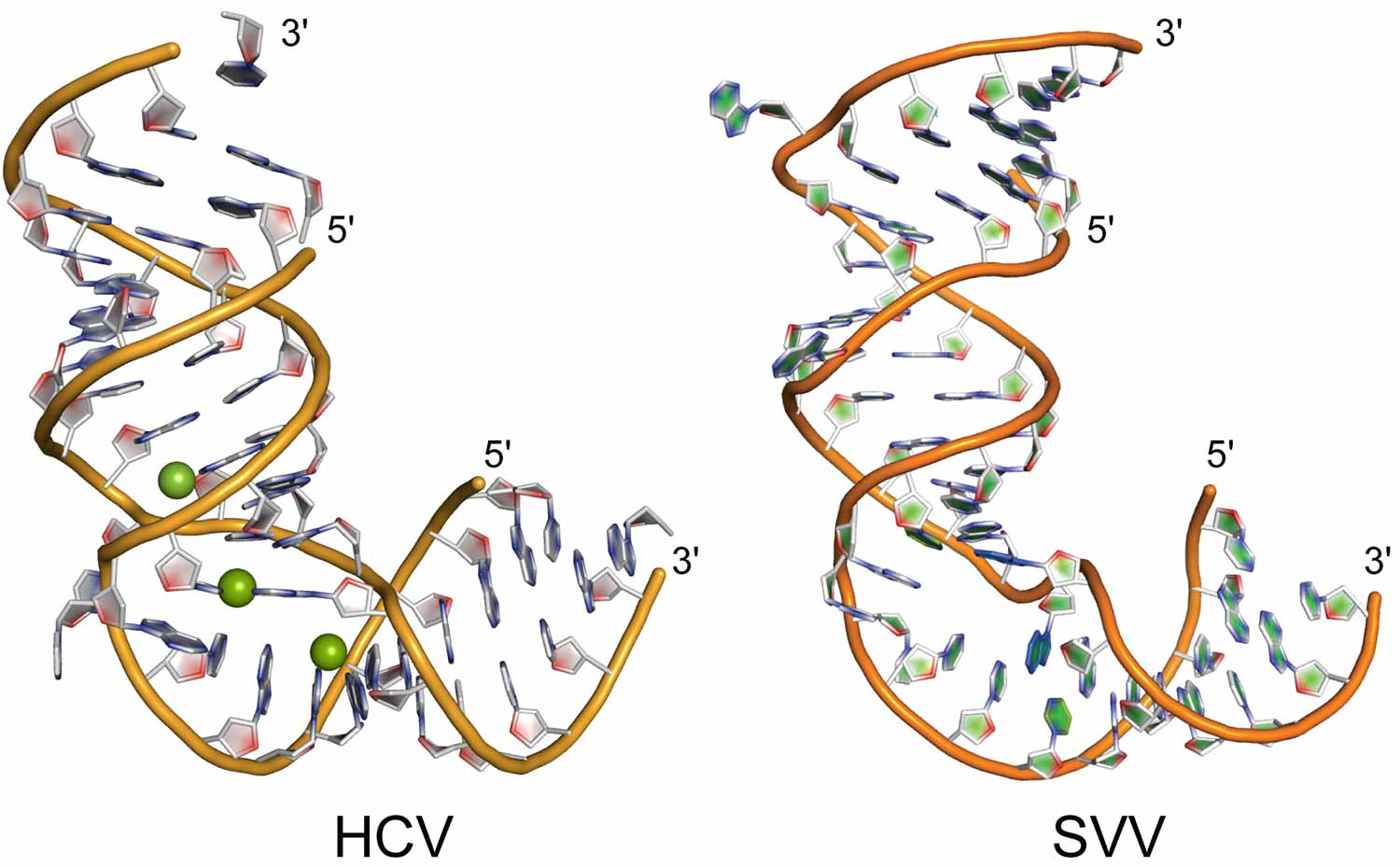
These on-off switches in viral genomes share a shape, but the details differ. RNA in hepatitis C virus (left) folds around metal ions; in Seneca Valley virus, the RNA bases cross over and stack. Credit: Hermann research group
A hinge in the RNA genome of the virus that causes hepatitis C works like a switch that can be flipped to prevent it from replicating in infected cells. Scientists have discovered that this shape is shared by several other viruses—among them one that kills cancer cells.
That’s Seneca Valley virus, which seems harmless to healthy human cells but lethal to cancer stem cells.
“Clearly we’d like to understand it better,” said Thomas Hermann, professor of chemistry and biochemistry at the University of California, San Diego.
Hermann’s research group has determined the molecular structure of this critical switch in the Seneca Valley virus and found that it matches the L-shaped switch in hepatitis C virus, which his group had previously described.
The similarity was a surprise.
“It wasn’t immediately apparent to us from the genome sequence alone,” said Mark Boerneke, a graduate student in chemistry and lead author of the report in the Proceedings of the National Academy of Sciences online the week of October 27. “This is a different virus that regulates protein production in a similar fashion.”
This kind of viral on-off switch is part of an IRES, for internal ribosome entry site. Ribosomes are the structures inside cells that make proteins. Lacking their own protein-making machinery, viruses use the IRES to hijack the ribosomes of cells they’ve infected to produce proteins they need to replicate.
Although switches for the two viruses share an overall L-shape, the sequences of the RNA that forms the Ls differ, as do the details of their structures. The L-shaped RNA in hepatitis C virus folds around metal ions; the switch in Seneca Valley virus has a cross-over motif of base pairs instead.
“Despite their overall similar shape, the details of the molecular interactions in the RNA switches are completely different,” Hermann said.
The researchers saw similar overall structures with differences in RNA sequence and molecular detail in three other viruses as well. The observation suggests that this L is a particularly effective shape, but the differences indicate that switches sharing the shape likely arose through separate evolutionary paths.
Yet they’re interchangeable switches. When Boerneke spliced the L-shaped RNA hinge from other viruses into the IRES of hepatitis C virus, it still functioned perfectly, but if he shifted the frame just a bit, with an extra base on one end and one missing from the other, the amount of viral proteins the modified virus could induce a host cell to make dropped by half.
Hermann’s group seeks small molecules potentially suitable as drugs that inhibit the workings of viruses that cause illness, like hepatitis C. They have previously identified one that straightened out the L-shaped switch in hepatitis C virus, preventing its RNA from inserting into the ribosome to even begin the process of making more copies of viral proteins.
One of the things that make viruses so difficult to combat is that they mutate rapidly. To get a handle on the variation in the hepatitis virus, Hermann’s group used data from genome sequencing of viruses collected from patients. These are viruses in the wild, with their rampant variation, rather than a uniform laboratory strain.
They found that the sequence that formed the L-shaped hinge in hepatitis C virus varied little. Geneticists call this “sequence conservation,” and it’s thought to be an indication that the sequence must be precise for the pathogen’s survival. The conservation of the L-shape and its ability to be straightened out by a small molecule suggests that a natural molecule fits into a pocket in the RNA.
In this case, one does: guanosine, one of the four bases found in the nucleic acids DNA and RNA.
“We found that the RNA switch is responsive to this guanosine trigger,” Hermann said.
Guanosine is simple, and so are these switches. They are much smaller—just 20 nucleotides long rather than hundreds—than previously described ”riboswitches” that respond to metabolites. The researchers think these viral RNA modules are about the simplest form a switch of this kind could take.
Although hepatitis C can now be cured with new drugs such as sofosbuvir and ledipasvir, these treatments are expensive. The 12-week daily dose prescribed for most patients would come to more than $80,000.
A better understanding of the RNA switch that shuts down viral replication could help chemists design better and perhaps less expensive inhibitors.
Additional authors include expert crystallographer Sergey Dibrov, Jing Gu, who contributed to the work as an undergraduate chemistry student researcher and Dr. David Wyles, a professor of medicine and a long-time collaborator of Hermann’s group. Wyles provided the virus, and Boerneke did some of the work in his lab. Hermann is a founder and director of the Center for Drug Discovery Innovation at UC San Diego in addition to his appointment in the Department of Chemistry and Biochemistry.
A grant from UC San Diego’s Academic Senate supported this work. The Biomolecule Crystallography Facility is supported by the National Institutes of Health and the Small Molecule NMR facility is supported by the National Science Foundation. Boerneke holds a Graduate Assistance in Areas of National Need (GAANN) fellowship from the U.S. Department of Education.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.


