
Varying Drug Levels in the Body Could Speed the Emergence of Drug-Resistant Bacteria
By:
- Susan Brown
Published Date
By:
- Susan Brown
Share This:
Article Content
Strains of bacteria able to resist multiple antibiotics pose a growing threat to public health, yet the means by which resistance quickly emerges aren’t well understood.
Scientists led by physics professor Terence Hwa at the University of California, San Diego, thought that the variety of environments in which bacteria encounter antibiotic drugs could play an important role. They have developed a mathematical model, published in the June 18 early online edition of the Proceedings of the National Academy of Sciences, that demonstrates how that would work.
Drug levels can vary widely between different organs and tissues in the human body, or between different individuals in a hospital. To account for that, their model considers a matrix of “compartments” with differing concentrations of a drug.
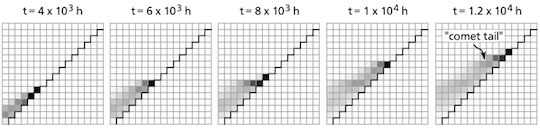
Snapshots from the model of evolving antibiotic resistance show a population of bacteria simultaneously expanding and adapting to antibiotic drugs, leaving a “comet tail” of less-resistant bacteria behind.
The bacteria in their model can move randomly from one compartment to the next. Their survival and rates of proliferation depend on the concentration of antibiotic within each compartment. And mutations that allow the bacteria to survive and thrive in environments with slightly higher concentrations were allowed to emerge randomly as well.
The system, designed to represent the varying environment of the human body, showed that drug-resistant mutants could evade competition by invading parts of the body, compartments in the model, with slightly higher drug concentrations where other bacteria fail to thrive.
When the process is repeated, the bacterial population can quickly adapt to components with much higher drug concentrations, with adaptation rates that would be very unlikely impossible in a uniform environment.
Although Hwa’s team created this model to study the evolution of antibiotic resistance, its formulation quite general. It could be applied to any example of adaptive expansion of an organism’s range, a general feature of biological systems that has allowed living things to populate every corner of the Earth, they write.
“Our mathematical model quantifies hypotheses, makes falsifiable predictions, and suggests experiments on this vast subject in which many words have been said but few quantitative statements can be found,” Hwa said. “The next step is quantitative experimentations which are being carried out in our lab and elsewhere.”
The iteration of quantitative prediction, experimental characterization, and model refinement characterizes quantitative biology, an emerging discipline that aims to fundamentally change biological research from discipline that is descriptive in nature to one that is quantitative and predictive. prerequisite for engineering and synthesis that promise to be the fruit of this century of biology.
Co-authors include post-doc Rutger Hermsen and graduate student Barret Deris, both members of Hwa’s research group. This work was supported by the Center for Theoretical Biological Physics (National Science Foundation) and the National Cancer Institute’s Physical Science-Oncology program. Deris holds a National Science Foundation Research Fellowship.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.


