
SDSC Supercomputer Modeling Reveals Acrobatics of CRISPR-Cas9 Technology
First step toward rational design of gene-splicing without “off-target” effects
By:
- Warren Froelich
Media Contact:
- Jan Zverina - jzverina@sdsc.edu
- Warren R. Froelich - froelich@sdsc.edu
Published Date
By:
- Warren Froelich
Share This:
Article Content
Cas9 Enzyme Approaching DNA, with the CRISPR Region in red. The CRISPR-Cas9 system is a transformative genome-editing tool. Here, Palermo et al. report the first biophysical study – based on multi-microsecond atomistic simulations – that details the conformational plasticity of the endonuclease Cas9 and its interplay with the nucleic acids. The study reveals a striking flexibility of the nuclease domain HNH as an essential element for its repositioning during the catalysis, with unprecedented key role of the non-target DNA strand in favoring the formation of a catalytically competent Cas9. These novel insights provide a foundation for structure engineering efforts for the rational design of new genome-engineering applications. Giulia Palermo, UC San Diego
A team led by researchers at the University of California San Diego has captured in step-by-step atomic detail the surgical editing of DNA strands by CRISPR-Cas9, the innovative gene-splicing technology that in recent years has transformed the field of genetic engineering.
Simulations performed by the Comet supercomputer at the San Diego Supercomputer Center (SDSC) at UC San Diego describe the “striking plasticity” of CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 and how it identifies, merges, and slices its target DNA strand. What’s more, the findings offer for the first hints at a key role played by the leftover non-target DNA strand, whose part in this biological cast of characters previously was unclear.
The goal of this study -- published in the September 8 issue of ACS Central Science, the new flagship journal of the American Chemical Society -- is to provide a foundation for the design of other novel, highly accurate genome-splicing technologies that don’t yield the “off-target” DNA breaks currently frustrating the potential of the current CRISPR-Cas9 system, particularly for clinical uses.
“CRISPR-Cas9 is not perfect since it can cause off-target effects or non-selective cleavage of DNA sequences, creating unwanted collateral damage,” said Giulia Palermo, a postdoctoral scholar with the UC San Diego Department of Pharmacology and lead author of the study.
“If we can design a very specific genome editing machinery, we can target the modification of genes controlling several diseases, including rare diseases and brain diseases, that are difficult to cure with available drugs,” added the study’s principal investigator J. Andrew McCammon, the Joseph E. Mayer Chair of Theoretical Chemistry, a Howard Hughes Medical Institute investigator, and Distinguished Professor of Pharmacology, all at UC San Diego.
“The rational design of more specific Cas9s, which are economically and environmentally friendly, and free from ethical issues, is our ultimate goal,” he said.
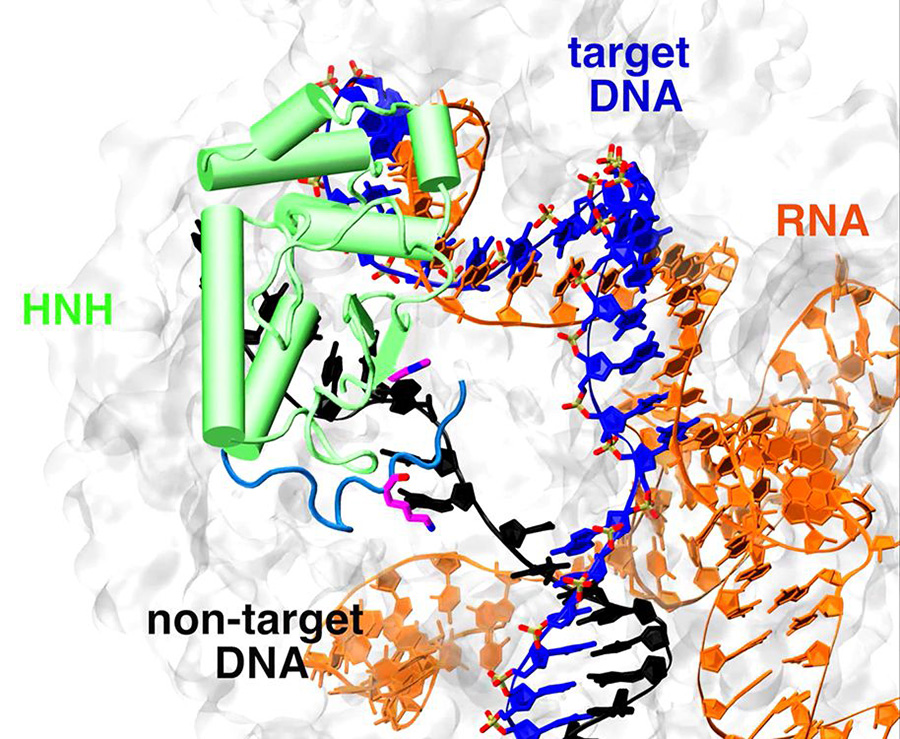
As its name implies, CRISPR-Cas9 is a dual entity with dual functions. The first consists of a short RNA guide molecule, part of which matches a target DNA sequence; the second is a Cas9 enzyme that recognizes and slices the DNA in a precise spot, whose location or address is post-marked by a nucleotide sequence called a protospacer adjacent motif, or PAM. The result is an RNA-DNA hybrid with a displaced non-target DNA strand.
Dubbed Science magazine’s “breakthrough of the year” in 2015, enthusiastic researchers around the world are just now scratching the surface of CRISPR-Cas9’s potential, with hopes of treating diseases through gene therapy, or driving advances in areas from crop engineering to the production of biofuels. What the technology ideally offers is specificity: the ability to target, edit, and insert new fragments of DNA sequences into the vast genome of the human and other species of animals and plants.
However, this transformative technology – known for the ease with which it can be programmed to cleave specific DNA targets – isn’t without its flaws. Studies have revealed that the RNA guide used to direct the cleaving enzyme to its target can sometimes go astray, landing on other DNA strands with similar but not identical sequences. The result is “off-target” mutations, severely limiting the technology’s vast array of potential applications, particular for human therapy.
Although extensive studies of the CRISPR-Cas9 systems, including X-ray crystallography and cryoelectron microscopy (cryoEM), have revealed detailed views of the system’s structure and biological activity, the dynamics of Cas9 and its step-by-step acrobatics with nucleic acids during its merger and cleavage of DNA have remained fuzzy at best.
To produce a motion picture-like view of this molecular interplay, UC San Diego researchers turned to the Comet supercomputer to perform atomistic molecular dynamics – a method that captures a more complete vision of the myriad shapes and conformations that a target protein molecule may go through – at petascale speeds (one quadrillion arithmetic calculations per second).
“Access to Comet, greatly facilitated by SDSC, was essential to completing this work in a reasonable timeframe,” said McCammon, also an SDSC Fellow and chemistry and biochemistry professor in UC San Diego’s Division of Physical Sciences. “The power of high-performance computing at the petascale-level and atomistic molecular dynamics simulations are needed to obtain key insights and relevant biophysical information that otherwise are inaccessible with currently available experimental techniques.”
The resulting simulations, performed over multi-microsecond timescales, revealed for the first time what the research team called the “remarkable” plasticity of the Cas9 system, and identified key factors underlying the myriad structural changes taking place during the merger and preparation for cleaving of its target DNA strand.
Of particular interest, the researchers were surprised to find that the leftover non-target DNA strand, whose role was generally considered unimportant, is actually a critical player in the system, serving as a type of starter key that triggers the final stage of the process.
“The motion and position of the non-target DNA strand triggers local conformational changes that result in a shift of an active domain site (HNH) of Cas9 towards the cleavage site on the target DNA for catalysis,” said McCammon, recently named the winner of the 2016-17 Joseph O. Hirschfelder Prize in Theoretical Chemistry, awarded by the Theoretical Chemistry Institute at the University of Wisconsin-Madison. “These molecular simulations strongly suggest the presence of non-target DNA as a key factor for the conformational activation of the HNH domain.”
Also participating the study, called “Striking plasticity of CRISPR-Cas9 and key role of non-target DNA, as revealed by molecular simulations”, were: Yinglong Miao, a research specialist with the Howard Hughes Medical Institute at UC San Diego and research scientist with the UC San Diego Department of Pharmacology; Ross C. Walker, associate research professor at SDSC, NVIDA Fellow, and adjunct associate professor in the Department of Chemistry and Biochemistry at UC San Diego; and Martin Jinek, currently an assistant professor at the University of Zurich who first discovered, in Jennifer Doudna’s lab at UC Berkeley, the ability of Cas9 to be programmed with single RNA strands for efficient DNA cleavage.
Funding for the study was provided by the Swiss National Science Foundation, in addition to grants to the McCammon lab from the National Institutes of Health, the National Science Foundation, and Howard Hughes Medical Institute; and research fellowships to Ross Walker from Intel and NVIDIA.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.


