
Quantitative Biology approach reveals importance of physical constraints on critical DNA interaction
Published Date
By:
- Susan Brown
Share This:
Article Content
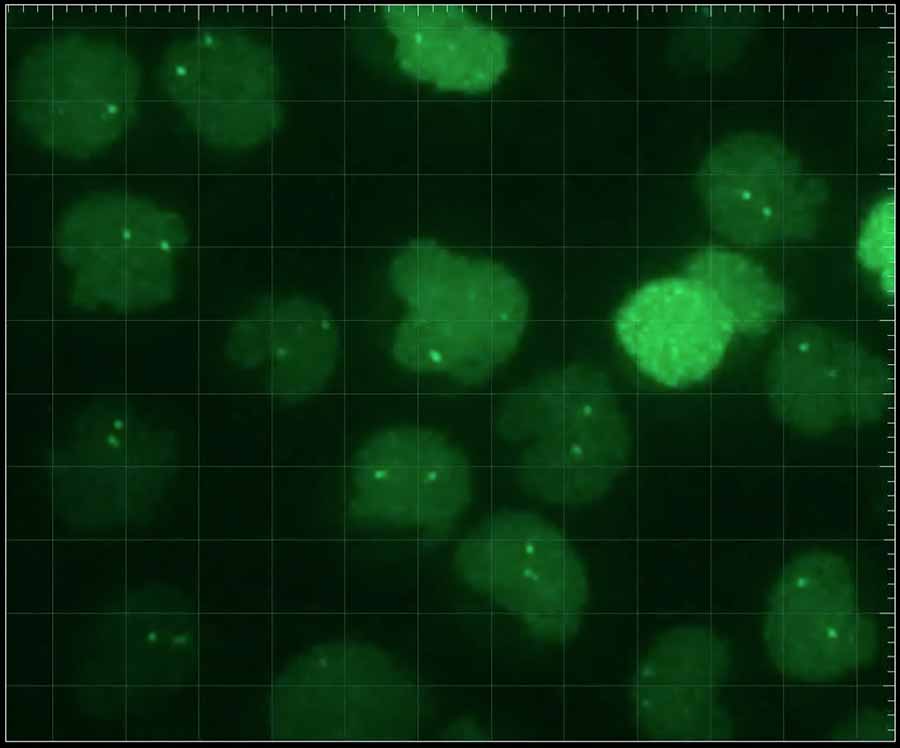
Fluorescent tags mark specific gene segements in living cells. Credit: Joseph Lucas
Our immune system copes with a multitude of threats using a mix-and-match system to create millions of different antibodies.
The white blood cells that produce these antibodies assemble their specific versions by selecting three gene segments from among multiple variants.
Joseph Lucas, a graduate student working with Cornelis Murre, a professor of biology at UC San Diego, tagged individual gene segments in live cells to track their movement in three dimensions.
To better understand what they observed, the biologists turned to Yaojun Zhang, a graduate student in physics and her advisor, Olga Dudko, a professor of physics at UC San Diego, who analyzed the movements. They recognized the patterns as ‘fractional Langevin motion.’
Rather than diffusing freely, like milk in black coffee, the movement of the elements is hindered by the mesh of proteins and nucleic acids inside the cell and the element’s attachment to stretchy chromatin fibers.
As a result of this viscoelastic quality of their environment, the gene segments likely bounce back and forth in a spring-like fashion until they meet and establish an interaction.
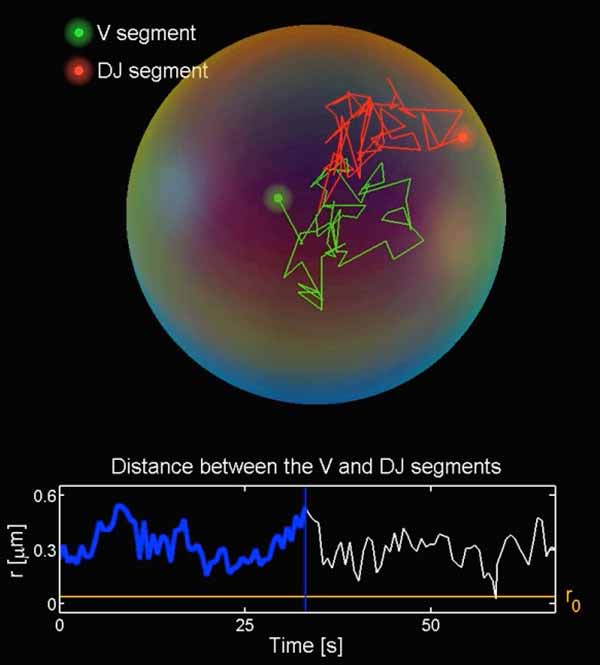
Numerical simulation of motion predicts distant genetic elements will meet quickly. Credit: Yaojun Zhang
Numerical simulations of the motions using fractional Langevin equations revealed how long it would take for individual genomic elements to first meet. Just seconds to minutes, hours at the outset, the team reported in a recent issue of Cell.
That’s quick, and the main factor seems to be that elements, though strung out separately along the chromatin, remain confined to a common genomic neighborhood.
We know from previous work that chromosomes fold into territories that rarely intermingle. These bundles of loops likely constrain movements and determine the timing of contacts between different genetic elements.
The importance of interactions between distant genetic elements goes beyond the production of antibodies. Regulatory elements control far flung coding elements to specify the suite of proteins made by particular types of cells. And the kind of genetic editing that creates highly specific antibodies sometimes goes awry causing breaks in DNA that can lead to cancer.
The National Institutes of Health and National Science Foundation supported this work.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.


