
Researchers Reveal 3D Structure of Cell’s Inflammation Sensor and its Inhibitors
Chemokine receptor study provides new drug development insights for autoimmune and inflammatory diseases
Published Date
By:
- Heather Buschman, PhD
Share This:
Article Content
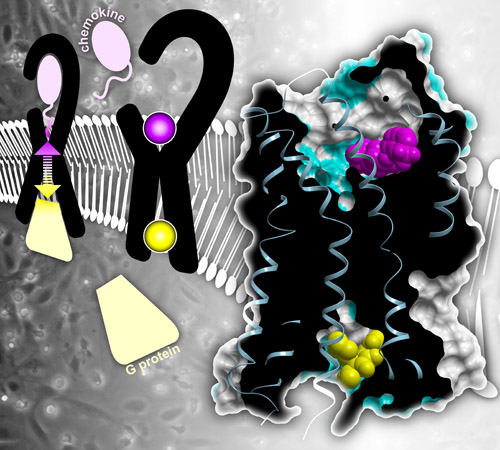
Right: Structure of CC chemokine receptor 2 (CCR2) bound to two inhibitors (pink and yellow). Left: Normally, chemokines outside the cell bind CCR2 and the receptor transmits that signal inside the cell via G proteins. Inhibitors block this signal transmission.
Inflammation is a good thing when it’s fighting off infection, but too much can lead to autoimmune diseases or cancer. In efforts to dampen inflammation, scientists have long been interested in CC chemokine receptor 2 (CCR2) — a protein that sits on the surface of immune cells like an antenna, sensing and transmitting inflammatory signals that spur cell movement toward sites of inflammation. Researchers at the Skaggs School of Pharmacy and Pharmaceutical Sciences at University of California San Diego have now determined the 3D structure of CCR2 simultaneously bound to two inhibitors. Understanding how these molecules fit together may better enable pharmaceutical companies to develop anti-inflammatory drugs that bind and inhibit CCR2 in a similar manner.
The study is published December 7 by Nature.
CCR2 and associated signaling molecules are known to play roles in a number of inflammatory and neurodegenerative diseases, including multiple sclerosis, asthma, diabetic nephropathy and cancer. Many drug companies have attempted to develop drugs that target CCR2, but none have yet made it to market.
“So far drugs that target CCR2 have consistently failed in clinical trials,” said Tracy Handel, PhD, professor in the Skaggs School of Pharmacy. “One of the biggest challenges is that, to work therapeutically, CCR2 needs to be turned ‘off’ and stay off completely, all of the time. We can’t afford ups and downs in its activity. To be effective, any small molecule drug that inhibits CCR2 would have to bind the receptor tightly and stay there. And that’s difficult to do.”
Handel led the study with Irina Kufareva, PhD, project scientist at Skaggs School of Pharmacy, and Laura Heitman, PhD, of Leiden University. The study’s first author is Yi Zheng, PhD, postdoctoral researcher also at Skaggs School of Pharmacy.
CCR2 spans the membrane of immune cells. Part of the receptor sticks outside the cell and part sticks inside. Inflammatory molecules called chemokines bind the external part of CCR2 and the receptor carries that signal to the inside of the cell. Inside the cell, CCR2 changes shape and binds other communication molecules, such as G proteins, triggering a cascade of activity. As a result, the immune cells move, following chemokine trails that lead them to places in the body where help is needed.
In this study, the researchers used a technique known as X-ray crystallography to determine the 3D structure of CCR2 with two molecules bound to it simultaneously — one at each end.
That’s a huge accomplishment because, Kufareva said, “Receptors that cross the cell membrane are notoriously hard to crystalize. To promote crystallization, we needed to alter the amino acid sequence of CCR2 to make the receptor molecules assemble in an orderly fashion. Otherwise, when taken out of the cell membrane, they tend to randomly clump together.”
Handel, Kufareva and team also discovered that the two small molecules binding CCR2 turn the receptor “off” by different but mutually reinforcing mechanisms. One of the small molecules binds the outside face of the receptor and blocks binding of the natural chemokines that normally turn the receptor "on.” The other small molecule binds the face of the receptor inside the cell, where the G protein normally binds, preventing inflammatory signal transmission. According to Handel, the latter binding site has never been seen before.
“It’s our hope that this new structure of CCR2 with two bound inhibitors will help optimize current and future drug discovery efforts,” Kufareva said.
Co-authors of this study also include: Ling Qin, Martin Gustavsson, Chunxia Zhao, Ruben Abagyan, UC San Diego; Natalia V. Ortiz Zacarías, Henk de Vries, Adriaan P. IJzerman, Leiden University; Gye Won Han, Vadim Cherezov, Raymond C. Stevens, University of Southern California; Marta Dabros, Robert Cherney, Percy Carter, Andrew Tebben, Briston-Myers Squibb Company; Dean Stamos, Vertex Pharmaceuticals.
This research was funded, in part, by the National Institutes of Health (R01GM071872, R01AI118985, R01GM117424, U54GM094618, U01GM094612, R21AI121918, R21AI122211, ACB-12002, AGM-12006).
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.


