
Scientists discover how Proteins interact along Metabolic Pathway
Research writes new chapter in the story of fatty acid biosynthesis and its potential for new antibiotics and renewable resources
Published Date
By:
- Cynthia Dillon
Share This:
Article Content
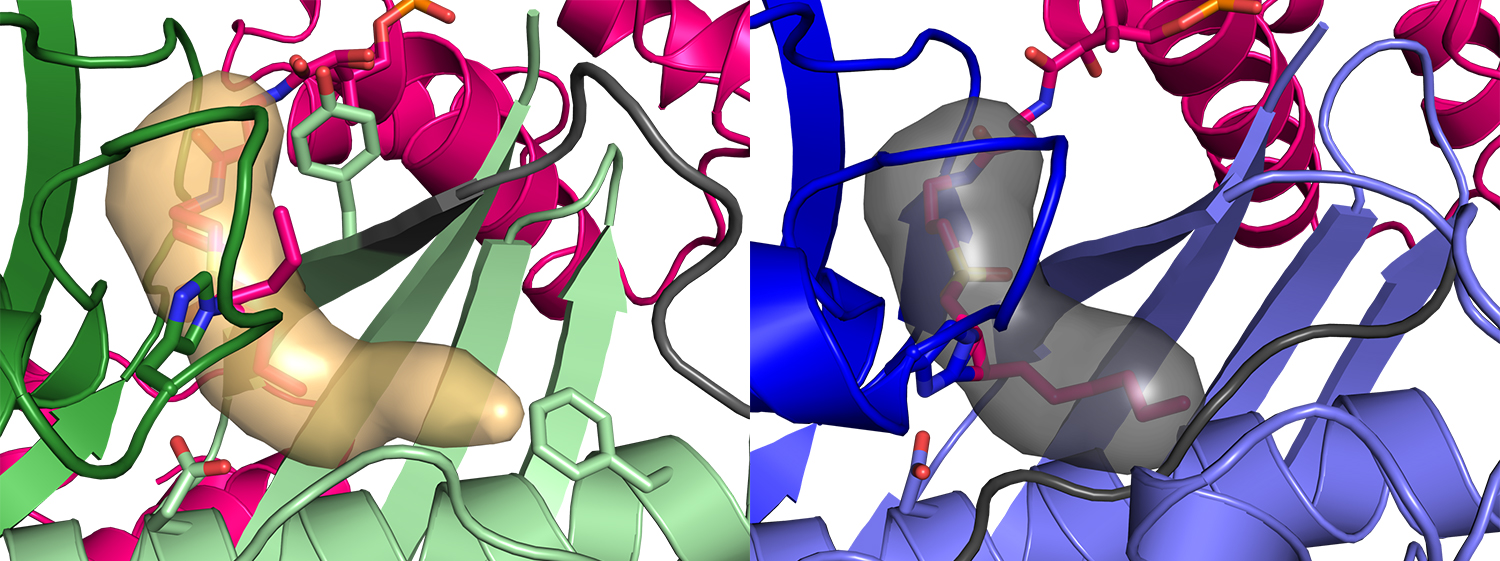
FabZ (green, left image) has a deep pocket (tan surface) for the fatty acid, but the fatty-acid pocket (gray surface, right image) has a different length and shape in FabA (blue). These differences and other details of the structures account for the different decisions of FabZ and FabA in making saturated (FabZ) vs. unsaturated (FabA) fatty acids. Images courtesy of Greg Dodge, University of Michigan
Life can depend upon the interaction of two proteins that come together along a metabolic pathway. There, they enable growth by creating fatty acids essential for ensuring that cell membranes form in a protective, fluid environment. While scientists have understood the basic enzymes involved in this fatty-acid biosynthesis, they have not always understood the many molecular transformations at play. In particular, they haven’t understood how the proteins interacted or what mechanistically allowed them to produce both saturated and unsaturated fats.
Now new research from scientists at the University of California San Diego and the University of Michigan has opened a new chapter in the story about what happens between two key metabolic enzymes. The findings, highlighted in a recent article published in Proceedings of the National Academy of Sciences, are setting chemical biologists on their own path to a new understanding of fatty acid biosynthesis.
“Nature likes to reuse the tools it created,” said UC San Diego’s Michael Burkart. “Not only does this work have relevance for the fundamental understanding of metabolism, but the findings can also be applied to the development of new antibiotics and for the synthetic biology of renewable fuels and materials.”

By applying Newtonian physics, Ashay Patel (left) and Michael Burkart (right), along with other researchers, could observe how metabolites behaved in the pocket, resulting in a time-resolved understanding of how proteins interact and catalyze specialized reactions. Photo by Michelle Fredricks, UC San Diego Physical Sciences
The study, initiated by former UC San Diego graduate student Kara Finzel and co-authored by Ashay Patel of UC San Diego and Greg Dodge of University of Michigan, fundamentally explains how bacteria—E. coli in this case—produce life’s critical saturated and unsaturated fatty acids.
Using chemical biology tools developed at UC San Diego, the authors worked with their mentors J. Andrew McCammon and Burkart, both leading researchers in the Department of Chemistry and Biochemistry at UC San Diego, and Janet Smith, a renowned X-ray crystallographer at the University of Michigan Life Sciences Institute, to closely examine a protein called AcpP and an enzyme called FabZ. The combined scientific expertise and methodology enabled the researchers to see the proteins in action.
Previous research revealed how AcpP is bound to FabZ’s partner enzyme, FabA. But researchers needed more information to determine precisely how these two enzymes produce two different fatty acids even though the enzymes themselves are very similar. By defining the structure of FabZ and its interactions with AcpP, these latest findings now demonstrate how the structures of the two enzymes enable them to perform different chemistry, resulting in either a saturated or an unsaturated fatty acid. In addition to defining the crystal structure, the researchers computed molecular dynamics simulations that enabled observations of the protein interaction and catalyzation.

One of the graduate students at work in the Burkart Lab at UC San Diego. Photo by Michelle Fredricks, UC San Diego Physical Sciences
The experiment’s results make clear how the enzyme called FabA uses a specialized pocket to rearrange atoms to retain the double bond that defines an unsaturated fat, and how this process is blocked in FabZ. Through the actions of these two enzymes, a single metabolic pathway can make both saturated and unsaturated fatty acids.
“This work explains how the decision is made, and then executed, to produce a saturated or unsaturated fat,” Smith noted. “It’s a fundamental advance in understanding how the structures and chemistry of these enzymes control the balance of fatty acids in the cell.”
This research was supported by the National Institutes of Health (NIH) (grant nos. GM31749, GM095970 and DK042303), the San Diego Supercomputer Center, the National Biomedical Computation Resource, the NIH National Cancer Institute (grant no. ACB-12002) and the National Institute of General Medical Sciences (grant no. AGM-12006).
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.


