SDSC Team Models Potential SARS-CoV2 Protease Inhibitors for COVID-19
‘Comet’ supercomputer helps illustrate possibilities for repurposing FDA-approved drugs
By:
- Kimberly Mann Bruch
Media Contact:
- Jan Zverina - jzverina@sdsc.edu
- Kimberly Mann Bruch - kbruch@ucsd.edu
Published Date
By:
- Kimberly Mann Bruch
Share This:
Article Content
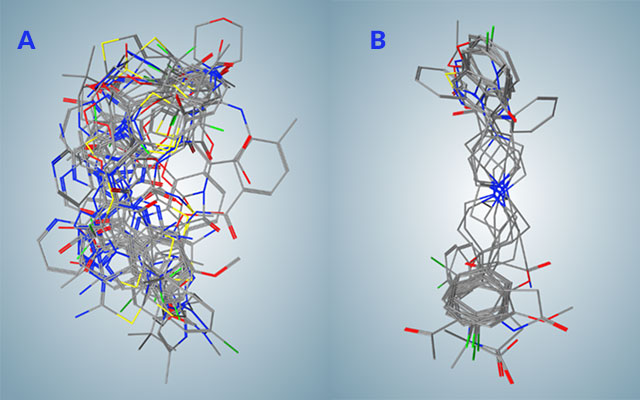
A research team at the San Diego Supercomputer Center (SDSC) on the UC San Diego campus recently created a pharmacophore model and conducted data mining of the database of drugs approved by the U.S. Federal Drug Administration (FDA) to find potential inhibitors of papain-like protease of SARS-CoV2, one of the main viral proteins responsible for COVID-19.
The study, recently published in PeerJ, found that among the drugs computationally selected were those currently used to inhibit HIV, Hepatitis C, and cytomesalovirus (CMV) as well as a set of drugs that have recently demonstrated positive effects on combating COVID-19.
The SDSC team included Igor Tsigelny, a researcher in structural biology, molecular modeling, and bioinformatics; Valentina Kouznetsova, who has expertise in protein modeling, metabolomics, biomarkers of diseases, and cancer diagnostics; Mark Miller (bioinformatics, phylogenetics eukaryotic cell biology, and metabolic engineering); and Mahidhar Tatineni, SDSC’s User Support Group Lead and a programmer analyst for high-performance computing. Tsigelny is also affiliated with the UC San Diego Moores Cancer Center.
SDSC’s Comet supercomputer was used for the computational docking of the selected drugs, covering a combined 4.3 million docking poses. After careful analysis, the researchers determined the most optimal compounds for simulated docking, which refers to the way drugs bind to papain-like protease.
“Once we completed the docking computations, we discovered that only some drugs had seemingly the best docking energies to bind viral protein that can help to fight COVID-19,” said Kouznetsova.
According to Tsigelny, some of the drugs selected by the pharmacophore search are already being experimentally tested for other viruses. He noted that that 11 compounds have been shown to be effective in treating other viruses - amodiaquine, chloroquine, sorafenib, dasatenib, hydroxychloroquine, bortezomib, topotecan, manidipine, lovastatin, cefitinib, and ritonavir.
Earlier this year, Kouznetsova and Tsigelny created a similar pharmacophore model that involved the data mining of the conformational database of FDA-approved drugs. With this earlier model, they identified 64 compounds as potential inhibitors of another SARS-CoV2 viral protein: main protease, also one of the main proteins responsible for COVID-19. Among the compounds selected then were two HIV protease inhibitors, two hepatitis C protease inhibitors, and three drugs that have already shown positive results in testing with COVID-19. Details of that study can be found here.
Support for this research included the San Diego Supercomputer Center. Comet is funded by the National Science Foundation (ACI #1341698). Mark Miller was supported by National Institutes of Health (R01 GM126463). Other authors received no funding for this work. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.