Scientists Discover a New Signaling Pathway and Design a Novel Drug for Liver Fibrosis
Health & Behavior
Membrane-associated proteins play a vital role in a variety of cellular processes, yet little is known about the membrane-association mechanism. Lipoprotein-associated phospholipase A2 (Lp-PLA2) is one such protein with an important role in cardiovascular health, but its mechanism of action on the phospholipid membrane was unknown. To address this, researchers at University of California San Diego School of Medicine used state-of-the-art experimental and computational tools to show exactly how the enzyme interacts with the membrane and extracts its specific substrates.
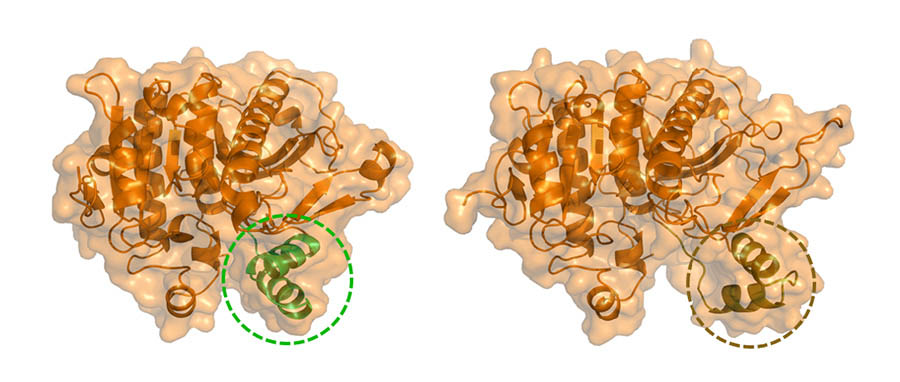
Lp-PLA2 in its “closed” conformation (left) and “open” conformation (right) once bound to the phospholipid monolayer.
The findings are publishing Jan. 3, 2022 in the online issue of Proceedings of the National Academy of Sciences.
Lp-PLA2 works on lipoproteins in the bloodstream, including common forms like low- and high-density lipoprotein (LDL and HDL). These lipoprotein particles are made up of a spherical layer of phospholipids surrounding a drop of fat and cholesterol esters. Over time, the phospholipids in this outer layer become oxidized, attracting free radicals and further oxidation, which contributes to plaque buildup and cardiovascular disease.
Lp-PLA2 extracts these oxidized phospholipids from the lipoprotein membrane and releases their fatty acids to be further metabolized. Understanding exactly how this process works creates new opportunities for therapeutics against cardiovascular disease.
“I am very pleased that we were able to go into much greater depth on how this enzyme works than ever before,” said Edward A. Dennis, PhD, senior author of the study and Distinguished Professor of Pharmacology, Chemistry and Biochemistry at UC San Diego School of Medicine. “Using the latest advances in lipidomics and computational molecular dynamics simulations, we got a picture which is worth a thousand words. We now have movies that show how this enzyme works at the atomic level, and that should help us figure out ways to activate or inactivate the enzyme as necessary for health.”
This advanced approach revealed a specific peptide region consisting of two alpha helices connected with a loop that acts as a gate to the enzyme’s active site. Typically, this gate is in a “closed” position, but when Lp-PLA2 binds to the phospholipid membrane, it undergoes an allosteric conformational change that opens the gate and increases the volume of the active site.
Dennis’ team, led by first author Varnavas D. Mouchlis, PhD, also showed which oxidized phospholipid substrates Lp-PLA2 has the greatest affinity for. They further identified a binding pocket distinct from known drug inhibitor binding pockets, which may serve as a new target for future therapeutic drugs.
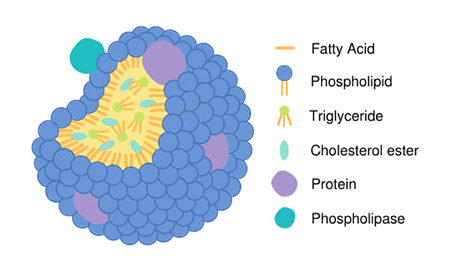
Schematic of a typical lipoprotein with a phospholipase bound to the outer phospholipid monolayer.
This study is the latest in a long line of work from the Dennis lab to develop a unifying theory on the function of phospholipases. The group had previously introduced this concept of membrane-facilitated allosteric regulation of PLA2 enzymes, but had until this point only studied enzymes that function on phospholipid bilayers (as seen on cells and intracellular organelles). This study confirmed that a similar mechanism could be used to facilitate phospholipase action on phospholipid monolayers, such as those on lipoproteins.
“PLA2 enzymes have all sorts of important functions in inflammation, digestion, brain health, and more, so it’s amazing to see this wide variety of enzymes all show a similar action strategy,” said Dennis. “We’ve been studying this superfamily of enzymes for almost 50 years, so to finally have this complete picture of how they work is really satisfying, and the whole field advances.”
Co-authors include: Daiki Hayashi, Alexis M. Vazquez, Jian Cao and J. Andrew McCammon, all at UC San Diego.
This research was funded, in part, by the National Institutes of Health (grants R01GM20501-44, R35GM139641 and R01GM31749).
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.