
Shedding Light on Cancer Diversity and Resistance
Published Date
By:
- Doug Ramsey
Share This:
Article Content
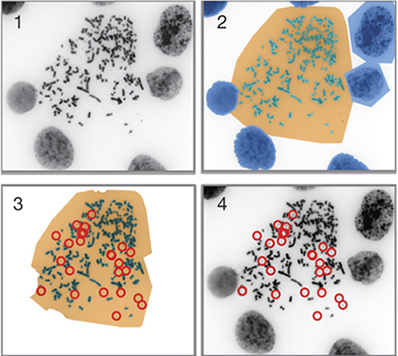
Schematic illustration of ECdetect (from Nature article linked below)
A paradigm-changing study by cancer researchers and computer scientists at the University of California San Diego and other institutions found that short fragments of circular DNA that encode cancer genes are far more common in cancer cells than previously believed. Published online in the journal Nature, the study* also concluded that the circular DNA probably plays a central role in generating the cellular diversity that makes advanced cancers so difficult to treat. The new findings are likely to change the way tumor evolution is understood by scientists and could ultimately lead to new ways to prevent and treat many malignancies.
The interdisciplinary team was led jointly by senior authors Paul Mischel from the university’s Ludwig Institute for Cancer Research, and bioinformatics expert Vineet Bafna, a professor of Computer Science and Engineering in UC San Diego’s Jacobs School of Engineering. "We've discovered something fundamental about how cancers diversify and evolve," said Mischel, a professor of pathology in the UC San Diego School of Medicine. "This is an essential rethinking about what goes wrong with genes in cancer."

Senior authors of the ecDNA paper in Nature: School of Medicine professor Paul Mischel (left) and Computer Science and Engineering professor Vineet Bafna
Graduate students working on the project analyzed cells from 17 different types of cancer to explore extrachromosomal DNA (ecDNA), which is so named because it is unassociated with chromosomes. They report that ecDNA is a key feature in nearly half of all types of tumors. The researchers also showed that ecDNA encodes multiple copies of cancer-driving genes.
“Their analysis also showed that ecDNA plays a far bigger role in the growth, diversity and drug resistance of cancer cells than the same genes housed on chromosomes in such tumors,” explained CSE’s Bafna, referring to the work done by two CSE Ph.D. students – Viraj Deshpande and Doruk Beyter – and first author Kristen Turner (a research scientist in the Ludwig Institute lab of Paul Mischel).
Turner acquired thousands of cytogenetic images, while CSE’s Beyter primarily did an unbiased computational analysis of those images to produce a systematic characterization regarding the extent and diversity of ecDNA across all 17 tumor subtypes.
“We also analyzed the genomic sequence by computational reconstruction of the structure of these matched extrachromosomal and intrachromosomal regions,” said CSE’s Vineet Bafna. “We found that extra- and intrachromosomal structures actually had a common origin.” The work on the origin of such structures was largely undertaken at UC San Diego by CSE grad student Viraj Deshpande.
To achieve their landmark results, the authors integrated several different technologies, including genomics, bioinformatics and classical cytogenetics, to detect, quantify, and analyze ecDNA. They found ecDNA in 40 percent of tumor cell lines but extremely rarely in normal cells. Further, when they looked specifically at patient-derived models of brain tumors, nearly 90 percent of these carried ecDNA.
Experimenting on tumor samples from patients, the researchers quantitatively modeled the findings and verified their model’s predictions. The studies revealed that tumors are more diverse -- or heterogeneous – when oncogenes are amplified on ecDNA instead of on chromosomes. This enabled them to achieve more rapidly and maintain high levels of cancer promoting genes.
Unlike chromosomes, ecDNA is parceled out randomly to daughter cells when a tumor cell divides. So any given cell in a tumor might have no ecDNA in its nucleus or be crammed to the proverbial gills with the stuff – and the greater the variation in their number, the greater the heterogeneity of cells in a tumor. It is this cellular diversity that makes tumors far more resistant to environmental challenges, most notably drug therapy.
“To understand the role of ecDNA formation in tumor evolution – in contrast to intrachromosomal amplification – we developed a mathematical model that allows us to contrast the two mechanisms,” said CSE’s Vineet Bafna. “Our models match the real data and help explain how ecDNA can provide tumor cells with a high diversity of ecDNA counts, allowing them to adapt more effectively to variable environmental conditions in a manner that maximizes tumor proliferation and survival.”
Bafna and his computer-science colleagues at UC San Diego were not part of a 2014 study led by Paul Mischel that inspired the latest research. In that previous study, Mischel and colleagues revealed that ecDNA plays a central role in the drug resistance of certain brain tumors. This came as a surprise because, for decades, cancer biologists had focused more onwhichgenes promote cancer rather thanwherethose genes are located.
Although a few cancer biologists in the 1960s described the presence of ecDNA in some tumor cells, they lacked the tools to quantify the presence of ecDNA, so the phenomenon had long been considered rare and inconsequential to the development of cancer. In the intervening years, this type of analysis was favored thanks to the evolution of genomic technologies.
"It occurred to us that… maybe ecDNA is a lot more common and consequential than anyone thought," noted Mischel. "Understanding how tumor cells evolve and how they increase the copy number and variability of their drivers is likely to yield some pretty important clues about the fundamental biology of cancer and how we might be able to target it." He and his team are now working to unearth the molecular mechanisms involved in the genesis and maintenance of ecDNA. They are also exploring how ecDNA levels change in response to changes in the tumor's internal environment.
The research was funded by Ludwig Cancer Research, the National Brain Tumor Society, the Ben and Catherine Ivy Foundation, the Ziering Family Foundation, the Susan G. Komen Foundation, the Leona M. and Harry B. Helmsley Charitable Trust, the Breast Cancer Research Foundation, the CureSearch for Children's Cancer, the California Institute for Regenerative Medicine, the NIH, and the NSF.
Geoffrey Wahl of the Salk Institute for Biological Studies’ Gene Expression Laboratory in La Jolla was also a co-senior author along with Bafna and Mischel. Other among the total of 21 contributors to the Nature paper included scientists from the Ludwig Institute, Sanford Burnham Prebys Medical Discovery Institute, UCLA Geffen School of Medicine, UC San Diego’s Moores Cancer Center, UC San Francisco Department of Pathology.
Share This:
You May Also Like
Stay in the Know
Keep up with all the latest from UC San Diego. Subscribe to the newsletter today.


